LLM Finetuning w/ SMILES-BERT
Motivation
In a previous blog post, I introduced the concept of semantic embedding search to improve the performance of a large language model (llm), and provided the source code implenetation for a conversational retrieval Q&A agent in LangChain. Today, I want to explore the alternative method of improving llm performance through finetuning on a downstream sequence classification task. This method is arguably more powerful than prompt tuning since finetuning can modify the model’s weights which parametrize its learned distribution over the dataset.
SMILES: language of chemical structure
In the last blog post on network analysis, I was working on the FooDB dataset which documents a large collection of compounds found in common foods. Each compound has a chemical structure represented by a data format known as SMILES, short for simplified molecular-input line-entry system. In formal language theory, SMILES is a context free language and can be modelled by a context free grammar (CFG) operating on finite sets of non-terminal, terminal, and start symbols. Alternatively, SMILES can also be interpreted as the string obtained from a depth-first tree traversal of a chemical graph. This graph is preprocessed into a traversable spanning tree by removing its hydrogen atoms and breaking its cycles. Numeric suffix labels are added to the symbols where the cycle was broken, and parentheses are used to indicate points of branching in the tree. Thus, the same chemical graph may have multiple valid SMILES representations, depending on our choice of where to break cycles, the starting atom for DFS, and the branching order. By adding constraints to some of these choices, algorithms have been developed to output canonical SMILES formats which can be converted back into its molecular graph.
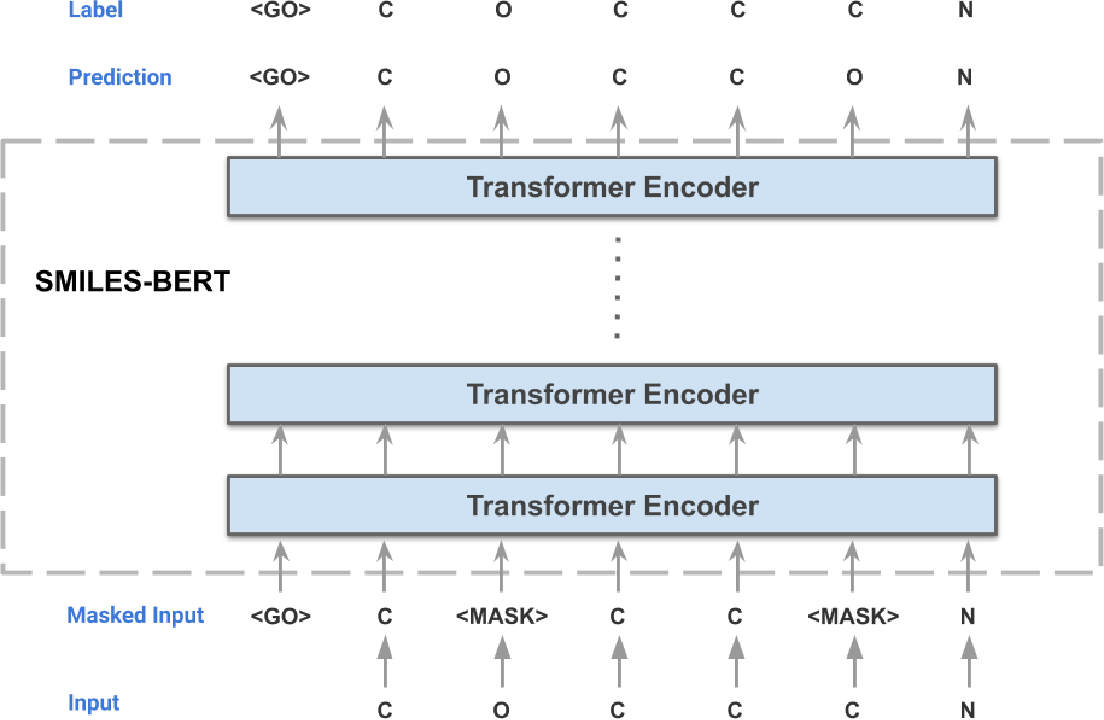
SMILES-BERT
Given the representational power of SMILES, attempts have been made to model this language through representation learning to capture the deep semantic knowledge contained in chemical graphs. Given that structure and function are intrinsically related to each other in biology, one would posit that learning the structural distribution of molecules could enable us to discover new relationships to function and other properties of interest. This leads us to SMILES-BERT, a BERT-like model trained on SMILES strings through a masked recovery task and finetuned on three downstream tasks of chemical property prediction.
Finetuning SMILES-BERT on FooDB compound library
Given my recent work on the FooDB compound library, which contains SMILES representations, I thought that it would be interesting to finetune the SMILES-BERT model on this dataset to further study the chemical properties tracked by the database.
To start, I decided to run the zero-shot SMILES-BERT model over the sequences and compress the embeddings using PCA to visualize the amount of inherent partitioning learned by the masked pretraining task. Here are the results when I color the projected embeddings by superclass, flavor, health effect, and ontology (function):
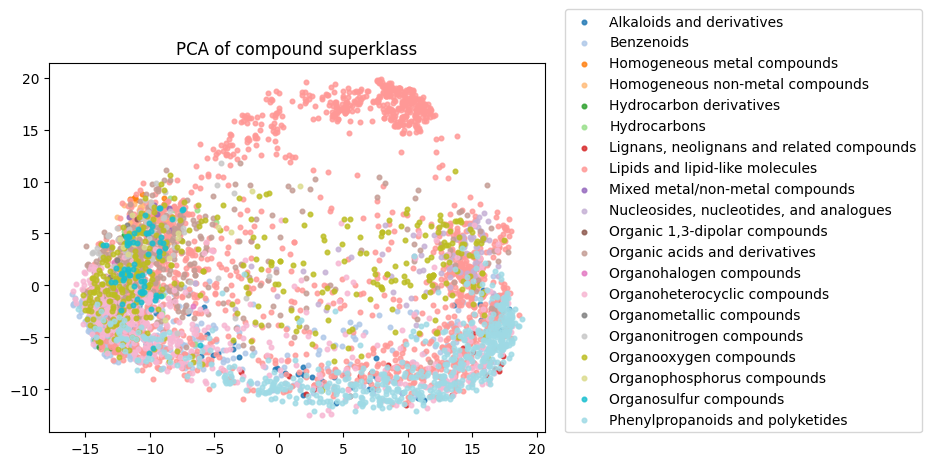
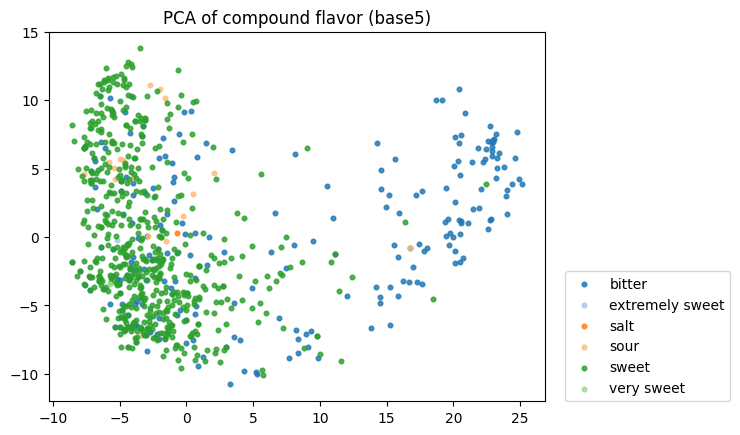
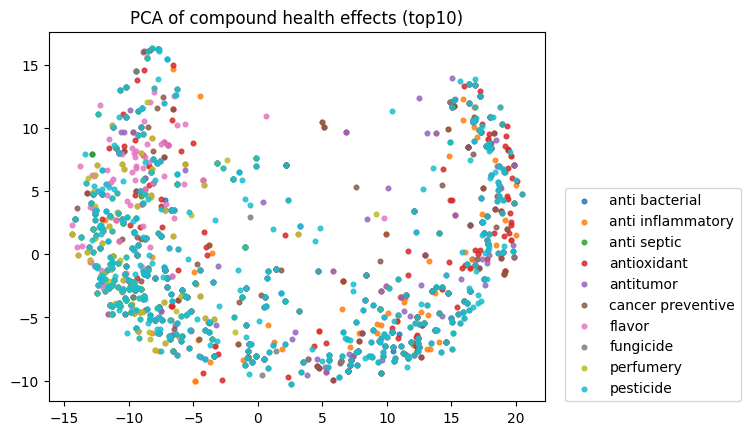
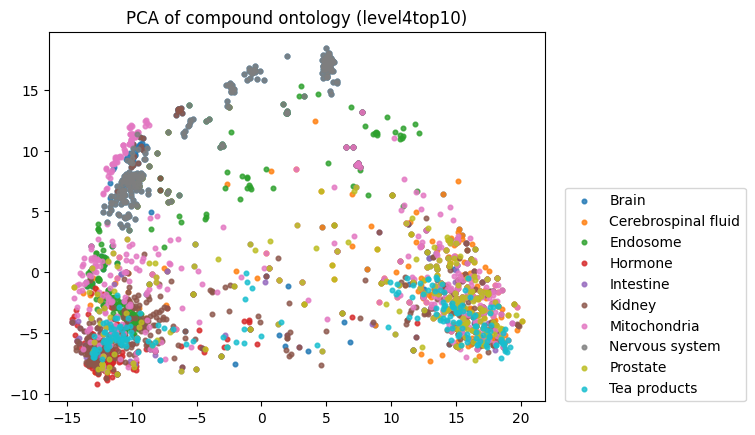
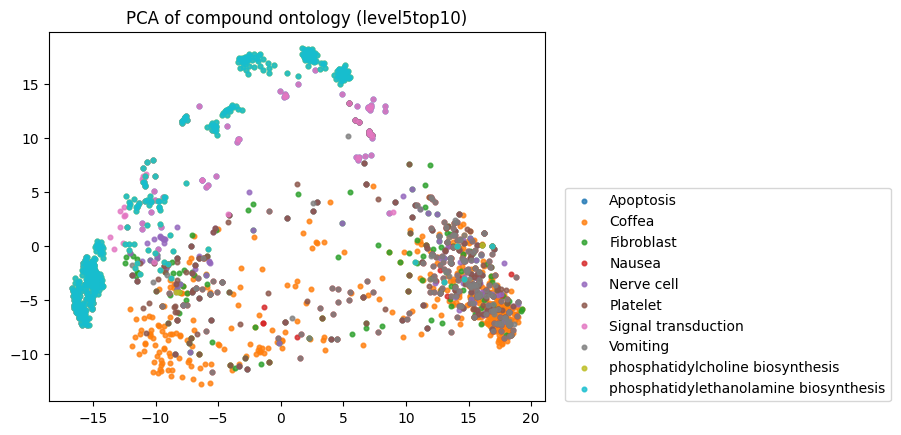
As you can see, SMILES-BERT clearly learns to differentiate some properties such as superclass and flavor, but struggles on some other categories like ontology.
Next, I wanted to actually finetune the model on these downstream classification tasks, then re-evaluate the model embedding projections on the held out test set to see if we can learn some non-linear transformations that actually enable us to better linearly separate these classes.
Notably, I used this model from the huggingface modelhub and separated my dataset into (80, 10, 10) training, validation, and testing splits. I ran the finetuning over 5 epochs with a batch size of 16, learning rate of 2e-5, with 0.01 weight decay, and AdamW optimizer. The complete code can be found in the following github repository.
Results
As expected, the finetuning yields better results on the categories that already had good embedding separation from the PCA results we saw earlier. From the training and eval loss curves plotted below, we can see that the model tends to overfit on the health effects and flavors categories.

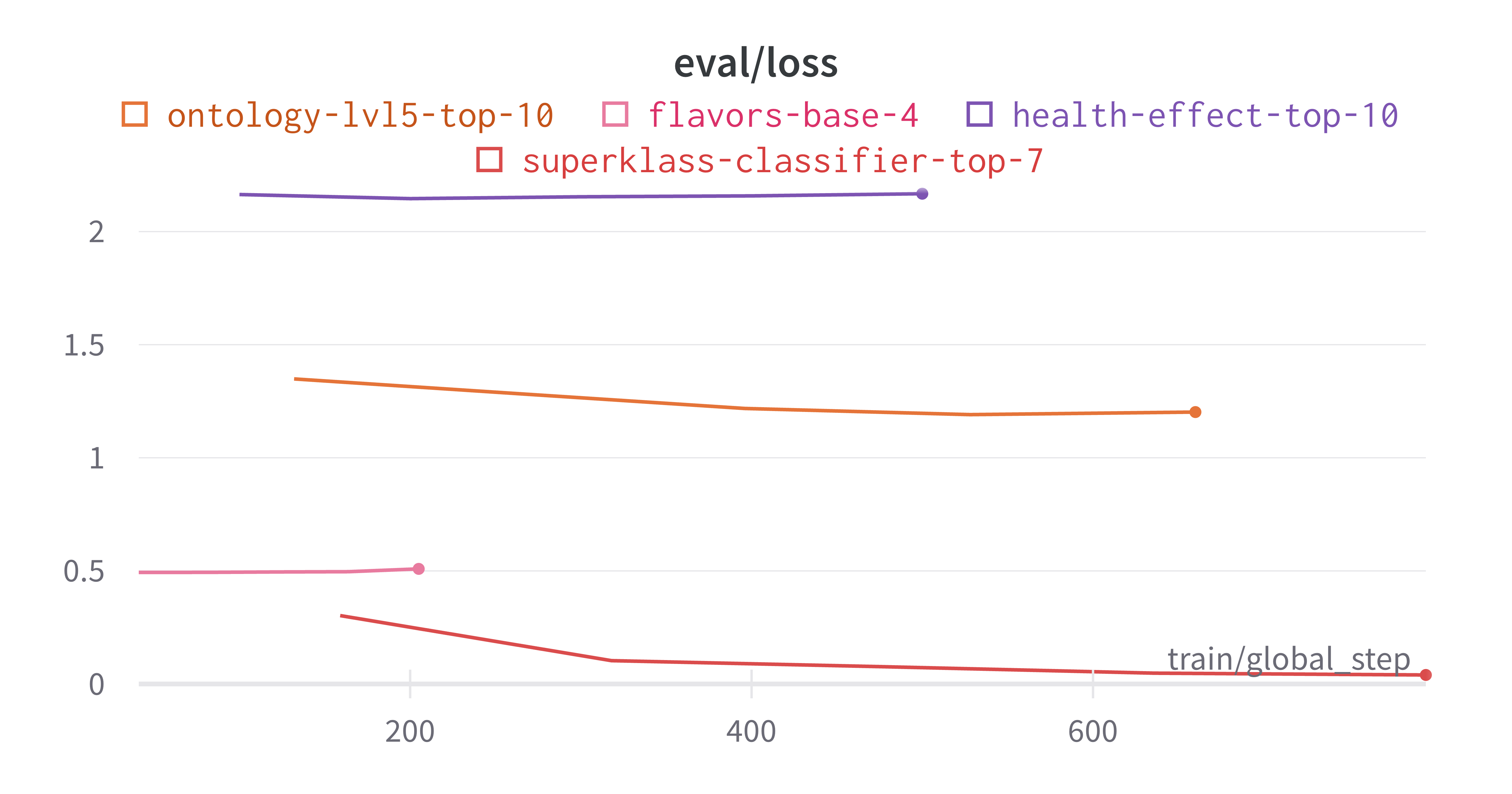
The best results I obtained were on the superklass category, and I think that this is because this categorization had the largest data to num_labels ratio, as well being fairly reliant on molecule structure which is captured by the SMILES strings. Here are the learning curves for this run where I obtained ~98% true accuracy on the held out test set.
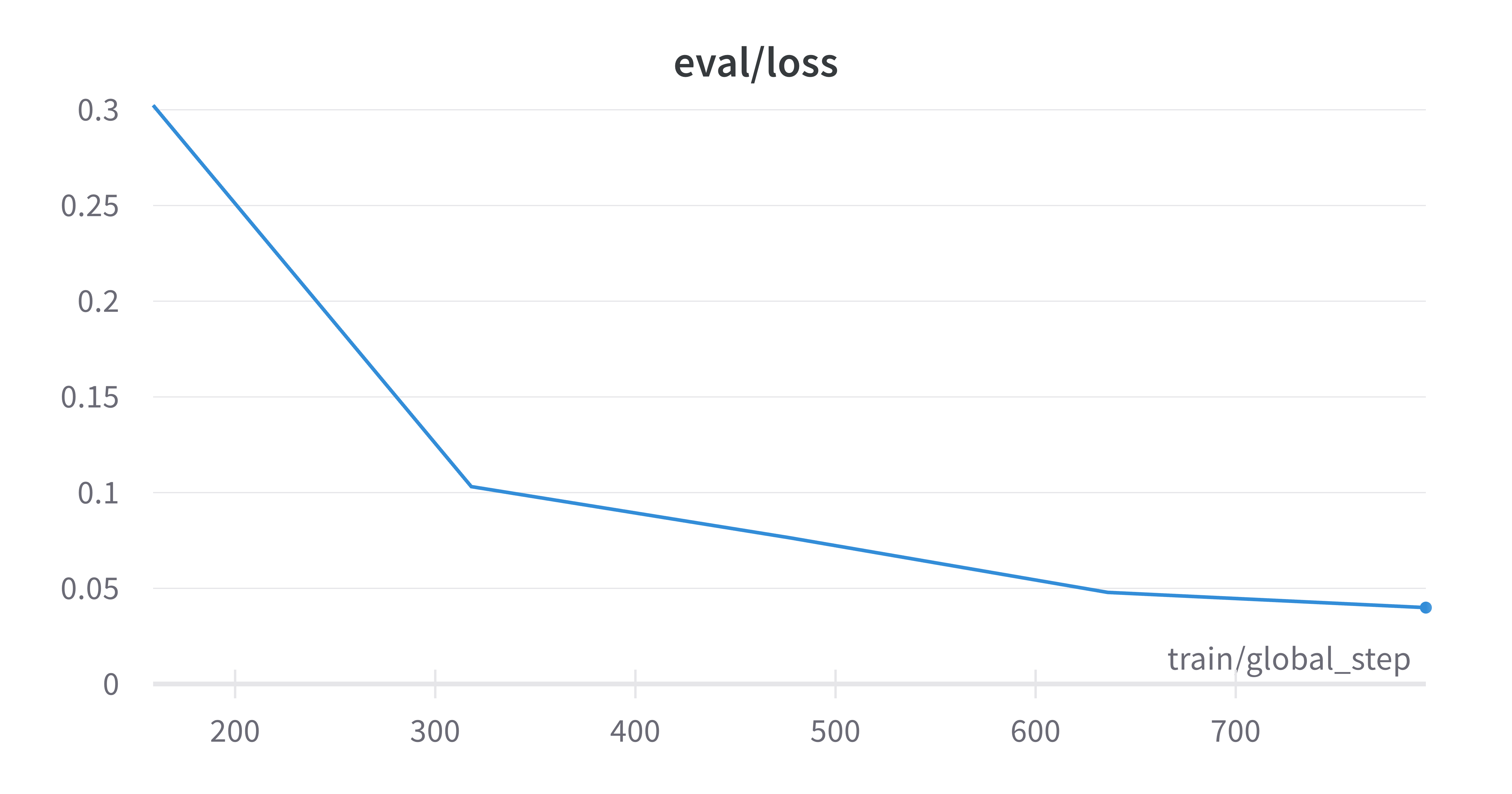

Finally, I reran this finetuned model on the raw SMILES strings to obtain embeddings and down-projected them using PCA. As you can see below, the model has clearly now learnt an embedding space that better separates the categories defined by the superklass label.
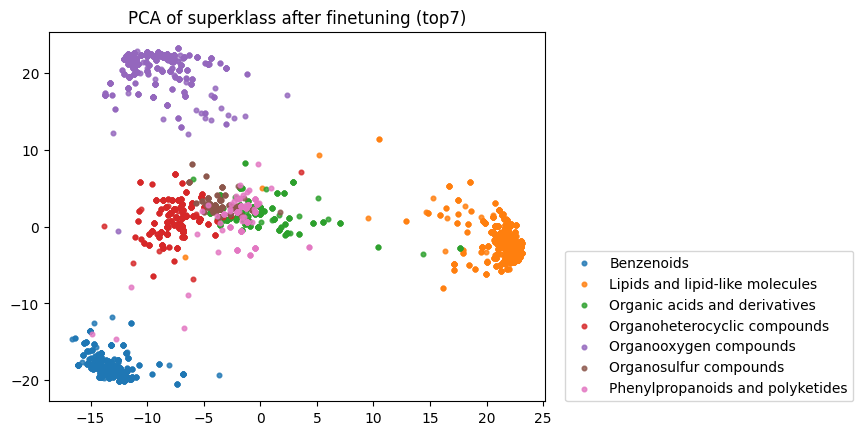
Compare this to the zero-shot embeddings below:

Enjoy Reading This Article?
Here are some more articles you might like to read next: